Artificial intelligence (AI) has been one of the most popular buzzwords in recent years. No matter what industry you’re working in, you more than likely have been inundated with the term “AI”—and rightfully so. AI has been redefining the way many organisations conduct their day-to-day operations, bringing improved work efficiency and saving resources simultaneously.
Machine learning (ML)—a subfield of AI which focuses on the use of data and algorithms to imitate the way humans learn, gradually improving its accuracy—has been a hot area for research and development. And perhaps no one better illustrates what ML is capable of than IBM’s Watson and Google’s DeepMind.
DeepMind is an ML system that uses algorithms based on deep neural networks and reinforcement learning to train massive datasets to be able to predict outcomes. It exploded onto the AI scene in 2016 when its AlphaGo program beat world champion Lee Sedol in four out of five games of Go—considered the world’s most complex board game by many. It achieved the feat—a decade earlier than expected—by combining “Monte-Carlo tree search with deep neural networks that have been trained by supervised learning, from human expert games, and by reinforcement learning from games of self-play,” according to Google.
Subsequently, many other programs were developed, such as AlphaZero—a general system that teaches itself from scratch how to master chess, shogi and Go—and Agent57—an ML agent capable of achieving scores above the human baseline on all 57 Atari 2600 games.
Learning by observing humans play at first, AlphaStar honed its skills by playing against itself, and successfully defeated professional StarCraft players 5-0! It acquired the abilities that traditional “built-in” gaming AIs lack—adaptability, improvisation and imagination—ingredients that make us human.
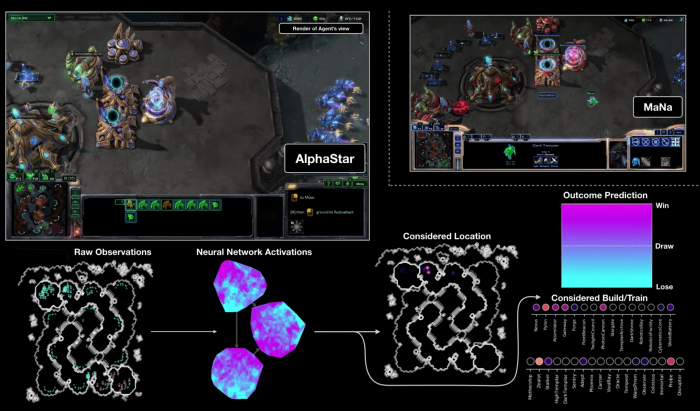
Screengrab from a YouTube video showing AlphaStar at work to defeat a professional StarCraft player. Outcome prediction shows that the AI system has victory within reach. Credits: Max Jaderberg/YouTube
While breaking record after record, DeepMind already has its mind set on something even more challenging. Something that researchers had been trying to solve for over five decades: predicting the structures of proteins.
Unfolding the mysteries of protein structure.
Proteins consist of strings of amino acids—the building blocks of everything in your body from muscle and hair to antibodies and enzymes. Nearly everything your body does, it does with proteins. And how a protein functions is determined by how it’s folded into its microscopic 3D shape, underpinning every biological process ever known to us.
Wonder how en egg “hard-boils”? It’s all just a simple tweak in protein chemistry.
“Structure is function” is an axiom of cellular biology. Understanding how ribbons of amino acids form the many complex twists and folds of a protein has significant health implications, from cancer to covid. As proteins are a primary target for many drugs and a crucial component in new therapeutics, unlocking their structures would accelerate the development of more effective treatments and therapies. And the key is AI.
AI key to unlocking the protein puzzle.
Predicting the structure of proteins, using cryo-electron microscopy or the “gold standard” x-ray crystallography, has historically been painstaking laboratory work. Entire PhDs have been dedicated to deciphering the folds of a single protein.
But all that changed when DeepMind introduced its AlphaFold AI system. Winning the CASP (Critical Assessment of Structure Prediction) Competition consecutively in 2018 (AlphaFold 1.0) and 2020 (AlphaFold 2.0), where computerised protein prediction methods are pitted against each other, the system was the first to accurately predict the structure of proteins that matched that of models produced in an experimental lab.
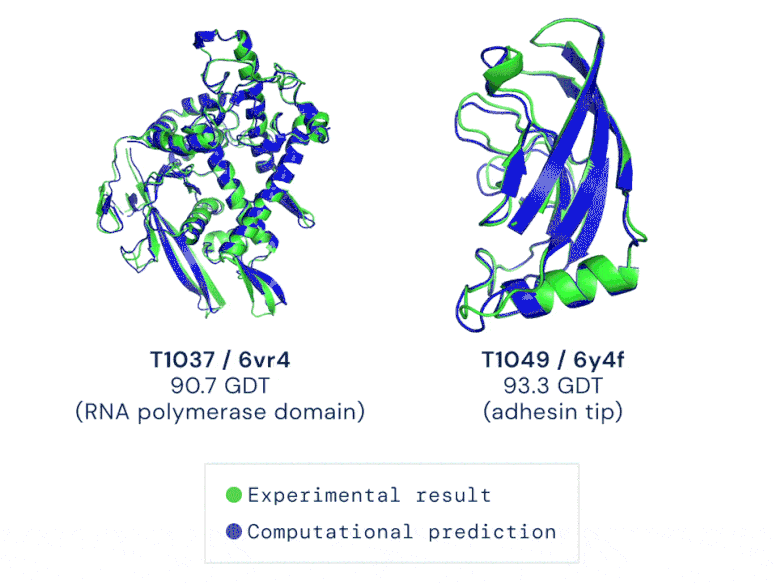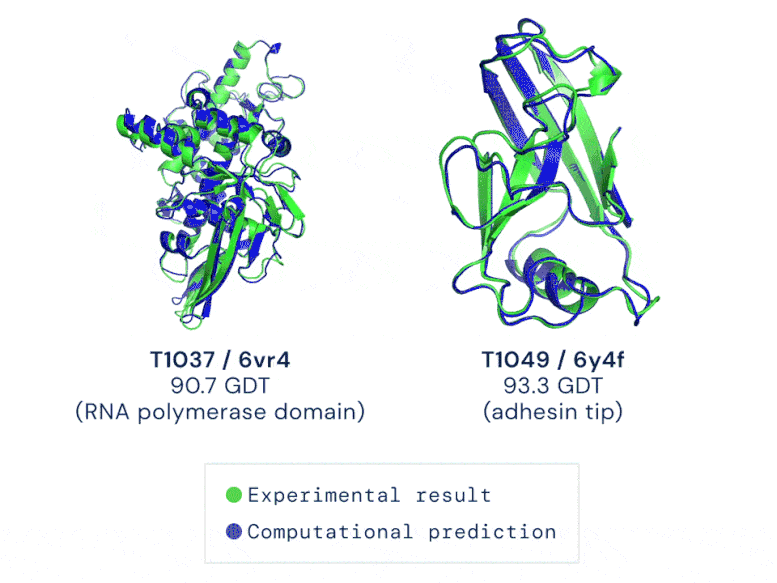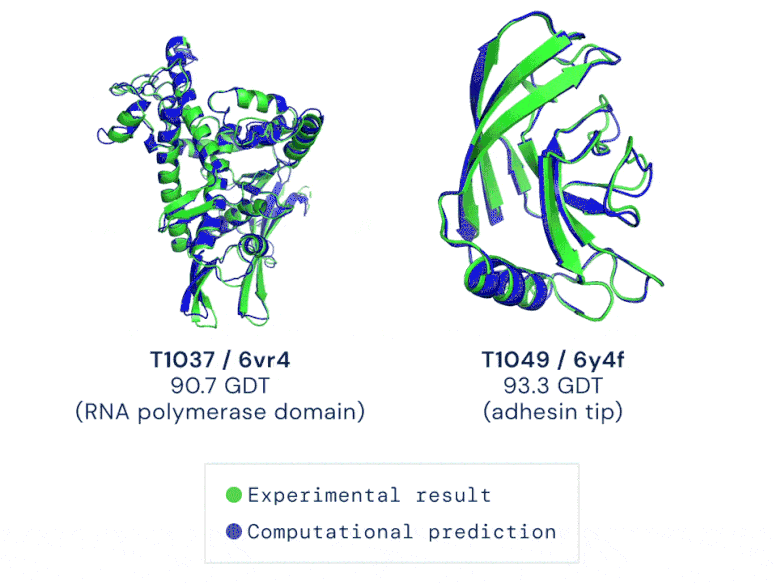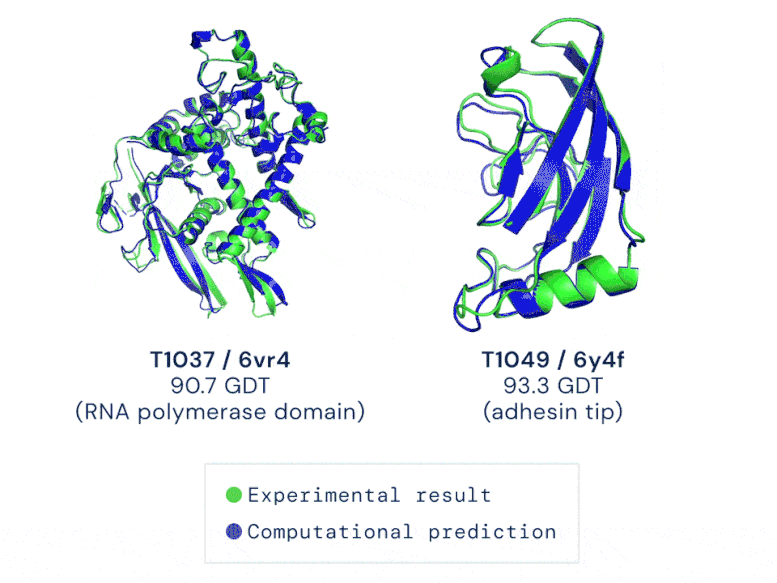
Two examples of protein targets in the free modelling category. AlphaFold predicts highly accurate structures measured against experimental results. GIF credit: DeepMind
The AI system—underpinned by an attention-based neural-network system— is so accurate that predictions are made with a margin of error of 1.6 angstroms (or 0.16 nanometres). That’s equivalent to roughly the width of an atom!
Recently, DeepMind published a paper detailing how the system works and released the source code. Now, a public database—openly available to the scientific community—is set up with over 800,000 entries of new protein structures. DeepMind also has plans to add more than 100 million—almost every single protein known to science—in the coming year.
DUG digs AlphaFold.
The AlphaFold program is installed on and optimised for our HPC system (connected to AARNet), ready for researchers to run any protein structure prediction on sequences not yet available on the AlphaFold Protein Structure Database.
Our HPC experts onboarded the AlphaFold source code, fine-tuned the associated workflow systems and ran tests based on terabytes of data to generate protein structures, ensuring that the program can fully leverage our state-of-the-art compute resources. It was also exciting to take AlphaFold for a spin on our newly installed NVIDIA Tesla A100 80GB GPUs!
Coupled with professional services such as code optimisation, algorithm development and on-demand software support, our experts allow you to divest yourself of time-consuming onboarding processes, enabling you to focus on what truly matters to you—the science.
Come and experience the brilliance of AlphaFold on our HPC, powered by DUG McCloud!