We are at the dawn of a new era of digital biology—where artificial intelligence (AI) and computational methods can help us better understand the models of important biological processes.
Recently the scientific community was stunned by a major breakthrough. DeepMind, a London-based AI company owned by Google, announced that its AI system AlphaFold had solved one of the the biggest mysteries in biology—predicting how a protein folds into a unique 3D shape.
From turning the tide on neglected diseases, to designing entirely new medical interventions, high performance computing (HPC)-enabled biology could have a huge impact on problems such as famine and disease, as well as opening up new avenues of scientific discovery.
Real-time tracking of pathogen evolution
Nextstrain is an open-source project that aims to harness the scientific and public health potential of pathogen genome data.
The project consists of a database of viral genomes, a bioinformatics pipeline for epidemiological analysis, and an interactive visualisation platform. Together these present a real-time view into the evolution and spread of a range of viral pathogens of high public health importance.
Through real-time tracking of pathogen evolution, scientists and clinicians can aid epidemiological understanding and improve outbreak response with effective public health measures and surveillance. Nextstrain is currently used by the Centers for Disease Control and Prevention, the national public health agency of the United States, to monitor SARS-CoV-2 genome sequences.
Mapping a virus’s march
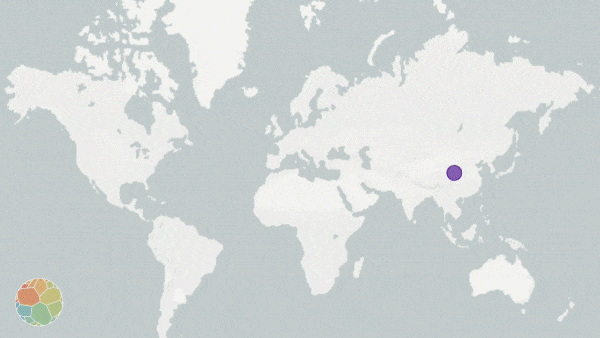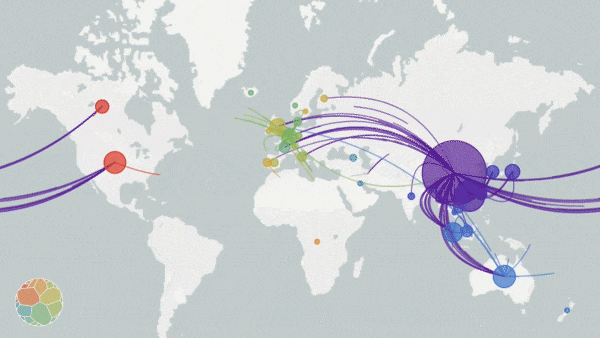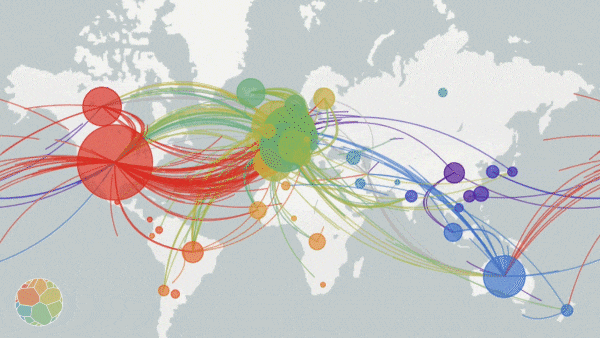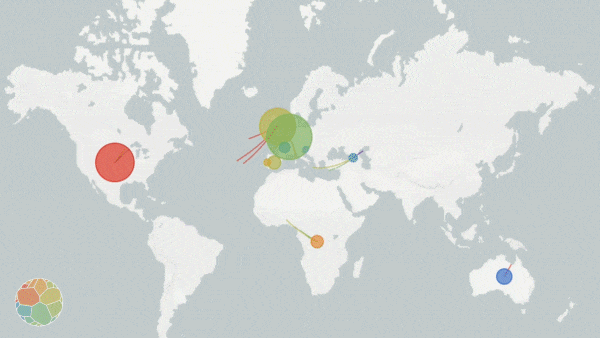
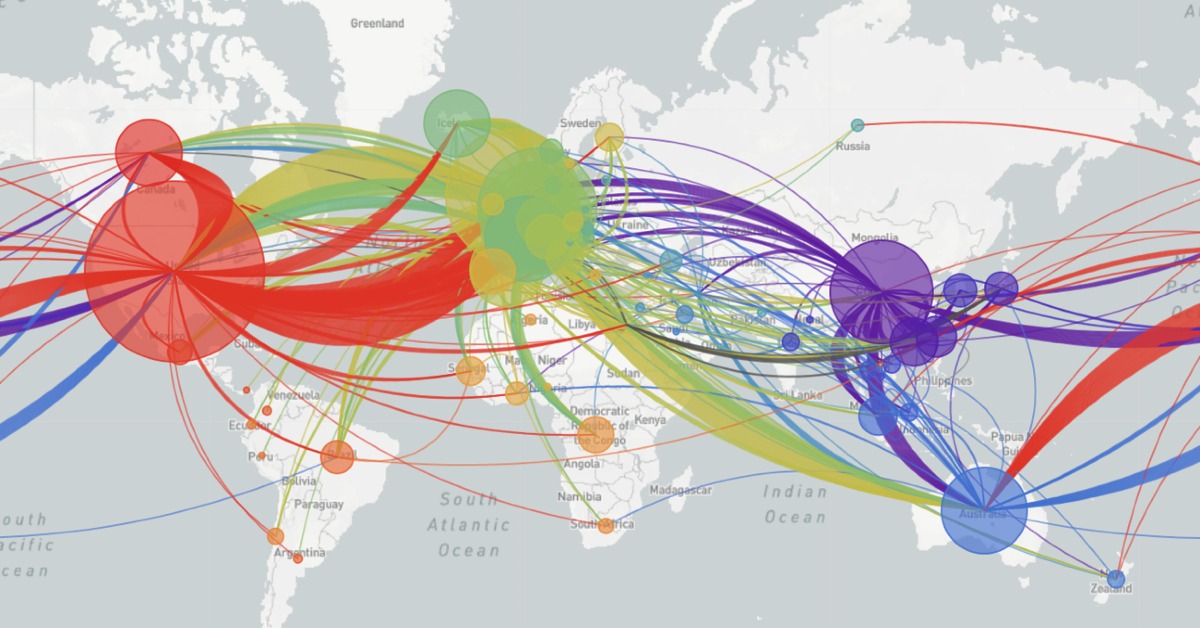
Animation on NextStrain.org showing COVID-19 strains circulating around the globe. The colours represent different variants of the SARS-CoV-2 virus whole the size of the circles represent the scale of the disease outbreak.
Originating from a single dot, lines start to extend. They stretch into Malaysia, India, Australia, and beyond. New colours paint up distant lands. And soon, a multi-coloured web has spun across the entire globe.
The pulsating map above tells the tale of the SARS-CoV-2 virus on the march—the story is written in the genetic code of the virus itself. Click here, scroll down to the world map, and press play to see an animated representation of the virus circulating throughout the globe.
When a virus infects a host, it churns out copies of itself. It’s a messy process, and some of those copies can have mutations, passed along when the virus infects others. As these mutations occur at a regular rate, they act like a ticking molecular clock. Researchers use that clock to figure out how and when those changes arose as viruses descended from a common origin.
Researchers use powerful technologies such as Nextstrain to quickly find mutations in samples from COVID-19 patients, building viral “family trees”, which let them trace the virus’s journey around the globe.
Nextstrain on high performance computing
Nextstrain can be used to track the real-time evolution of diseases, from COVID-19 and monkeypox to the seasonal flu. It’s made up of Augur, a bioinformatics toolkit for phylogenetic analysis, and Auspice, an open-source interactive tool for visualising phylogenomic data.
Nextstrain installed on and optimised for our HPC system—connected to AARNet—ready for researchers to run phylogenetic analysis and visualisation for local modelling, or for submission to the Nextstrain website for global tracking. With thousands of machines ready to run at DUG, you’re empowered to greatly reduce your analysis time, improving the turnaround for your time-critical research.
Drop us an email at [email protected] to find out how we can help accelerate your bioinformatics projects today.